Example PD – Advanced data loading with pandas and ROOT
Aims
Instructions
Pandas is an open source, BSD-licensed library providing high-performance, easy-to-use data structures and data analysis tools for the Python programming language:
It provides a DataFrame class, which is a useful tool to organise
structured data:
from remu import binning
from remu import plotting
import numpy as np
import pandas as pd
pd.set_option('display.max_rows', 10)
px = np.random.randn(1000)*20
py = np.random.randn(1000)*20
pz = np.random.randn(1000)*20
df = pd.DataFrame({'px': px, 'py': py, 'pz': pz})
print(df)
px py pz
0 -2.840325 22.108657 -0.516603
1 1.021440 41.311321 9.110285
2 -2.311842 7.761168 34.076248
3 31.893287 -12.497031 -9.125932
4 12.025659 3.006832 -18.585591
.. ... ... ...
995 -3.413603 21.201968 -21.732508
996 30.549099 19.142792 30.115672
997 5.267751 20.139826 11.095047
998 15.030364 -7.931964 4.888165
999 -8.671663 17.492177 21.862662
[1000 rows x 3 columns]
ReMU supports DataFrame objects as inputs for all
fill methods:
with open("muon-binning.yml", 'r') as f:
muon_binning = binning.yaml.full_load(f)
muon_binning.fill(df)
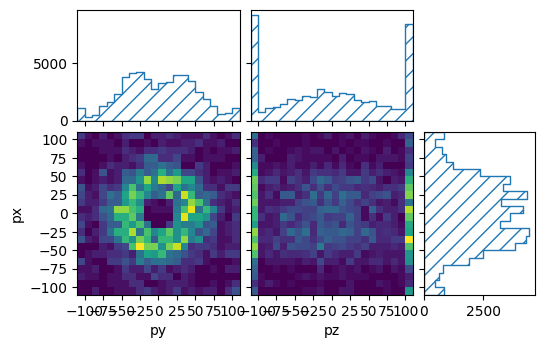
pltr = plotting.get_plotter(muon_binning, ['py','pz'], ['px'])
pltr.plot_values()
pltr.savefig("pandas.png")
This way, ReMU supports the same input file formats as the pandas library, e.g. CSV, JSON, HDF5, SQL, etc..
Using the uproot library, pandas can also be used to load ROOT files:
https://github.com/scikit-hep/uproot5
The ROOT framework is the de-facto standard for data analysis in high energy particle physics:
Uproot does not need the actual ROOT framework to be installed to work. It
can convert a flat ROOT TTree directly into a usable pandas
DataFrame:
import uproot
flat_tree = uproot.open("Zmumu.root")['events']
print(flat_tree.keys())
['Type', 'Run', 'Event', 'E1', 'px1', 'py1', 'pz1', 'pt1', 'eta1', 'phi1', 'Q1', 'E2', 'px2', 'py2', 'pz2', 'pt2', 'eta2', 'phi2', 'Q2', 'M']
df = flat_tree.arrays(library="pd")
print(df)
Type Run Event E1 px1 ... pt2 eta2 phi2 Q2 M
0 GT 148031 10507008 82.201866 -41.195288 ... 38.8311 -1.051390 -0.440873 -1 82.462692
1 TT 148031 10507008 62.344929 35.118050 ... 44.7322 -1.217690 2.741260 1 83.626204
2 GT 148031 10507008 62.344929 35.118050 ... 44.7322 -1.217690 2.741260 1 83.308465
3 GG 148031 10507008 60.621875 34.144437 ... 44.7322 -1.217690 2.741260 1 82.149373
4 GT 148031 105238546 41.826389 22.783582 ... 21.8913 1.444340 -2.707650 -1 90.469123
... ... ... ... ... ... ... ... ... ... .. ...
2299 GG 148029 99768888 32.701650 19.054651 ... 22.8145 -0.645971 -2.404430 -1 60.047138
2300 GT 148029 99991333 168.780121 -68.041915 ... 32.3997 -1.570440 0.037027 1 96.125376
2301 TT 148029 99991333 81.270136 32.377492 ... 72.8781 -1.482700 -2.775240 -1 95.965480
2302 GT 148029 99991333 81.270136 32.377492 ... 72.8781 -1.482700 -2.775240 -1 96.495944
2303 GG 148029 99991333 81.566217 32.485394 ... 72.8781 -1.482700 -2.775240 -1 96.656728
[2304 rows x 20 columns]
muon_binning.reset()
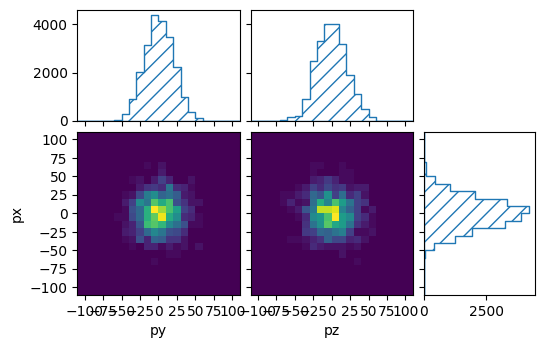
muon_binning.fill(df, rename={'px1': 'px', 'py1': 'py', 'pz1': 'pz'})
pltr = plotting.get_plotter(muon_binning, ['py','pz'], ['px'])
pltr.plot_values()
pltr.savefig("flat_muons.png")
ReMU expects exactly one row per event. If the root file is not flat, but has a more complicated structure, it must be converted first. For example, let us take a look at a file where each event has varying numbers of reconstructed particles:
structured_tree = uproot.open("HZZ.root")['events']
print(structured_tree.keys())
['NJet', 'Jet_Px', 'Jet_Py', 'Jet_Pz', 'Jet_E', 'Jet_btag', 'Jet_ID', 'NMuon', 'Muon_Px', 'Muon_Py', 'Muon_Pz', 'Muon_E', 'Muon_Charge', 'Muon_Iso', 'NElectron', 'Electron_Px', 'Electron_Py', 'Electron_Pz', 'Electron_E', 'Electron_Charge', 'Electron_Iso', 'NPhoton', 'Photon_Px', 'Photon_Py', 'Photon_Pz', 'Photon_E', 'Photon_Iso', 'MET_px', 'MET_py', 'MChadronicBottom_px', 'MChadronicBottom_py', 'MChadronicBottom_pz', 'MCleptonicBottom_px', 'MCleptonicBottom_py', 'MCleptonicBottom_pz', 'MChadronicWDecayQuark_px', 'MChadronicWDecayQuark_py', 'MChadronicWDecayQuark_pz', 'MChadronicWDecayQuarkBar_px', 'MChadronicWDecayQuarkBar_py', 'MChadronicWDecayQuarkBar_pz', 'MClepton_px', 'MClepton_py', 'MClepton_pz', 'MCleptonPDGid', 'MCneutrino_px', 'MCneutrino_py', 'MCneutrino_pz', 'NPrimaryVertices', 'triggerIsoMu24', 'EventWeight']
df = structured_tree.arrays(["NMuon", "Muon_Px", "Muon_Py", "Muon_Pz"], library='pd')
print(df)
NMuon ... Muon_Pz
0 2 ... [-8.16079330444336, -11.307581901550293]
1 1 ... [20.199968338012695]
2 2 ... [11.168285369873047, 36.96519088745117]
3 2 ... [403.84844970703125, 335.0942077636719]
4 2 ... [-89.69573211669922, 20.115053176879883]
... ... ... ...
2416 1 ... [61.715789794921875]
2417 1 ... [160.8179168701172]
2418 1 ... [-52.66374969482422]
2419 1 ... [162.1763153076172]
2420 1 ... [54.71943664550781]
[2421 rows x 4 columns]
This kind of data frame with “lists” as cell elements can be inconvenient to handle. But we can flatten it using the power of the awkward:
import awkward as ak
arr = structured_tree.arrays(["NMuon", "Muon_Px", "Muon_Py", "Muon_Pz"])
df = ak.to_dataframe(arr)
print(df)
NMuon Muon_Px Muon_Py Muon_Pz
entry subentry
0 0 2 -52.899456 -11.654672 -8.160793
1 2 37.737782 0.693474 -11.307582
1 0 1 -0.816459 -24.404259 20.199968
2 0 2 48.987831 -21.723139 11.168285
1 2 0.827567 29.800508 36.965191
... ... ... ... ...
2416 0 1 -39.285824 -14.607491 61.715790
2417 0 1 35.067146 -14.150043 160.817917
2418 0 1 -29.756786 -15.303859 -52.663750
2419 0 1 1.141870 63.609570 162.176315
2420 0 1 23.913206 -35.665077 54.719437
[3825 rows x 4 columns]
This double-index structure is still not suitable as input for ReMU, though. We can select only the first muon in each event, to get the required “one event per row” structure:
idx = pd.IndexSlice
df = df.loc[idx[:,0], :]
print(df)
NMuon Muon_Px Muon_Py Muon_Pz
entry subentry
0 0 2 -52.899456 -11.654672 -8.160793
1 0 1 -0.816459 -24.404259 20.199968
2 0 2 48.987831 -21.723139 11.168285
3 0 2 22.088331 -85.835464 403.848450
4 0 2 45.171322 67.248787 -89.695732
... ... ... ... ...
2416 0 1 -39.285824 -14.607491 61.715790
2417 0 1 35.067146 -14.150043 160.817917
2418 0 1 -29.756786 -15.303859 -52.663750
2419 0 1 1.141870 63.609570 162.176315
2420 0 1 23.913206 -35.665077 54.719437
[2362 rows x 4 columns]
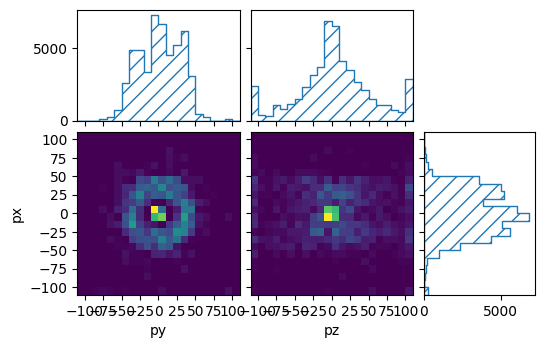
muon_binning.reset()
muon_binning.fill(df, rename={'Muon_Px': 'px', 'Muon_Py': 'py', 'Muon_Pz': 'pz'})
pltr = plotting.get_plotter(muon_binning, ['py','pz'], ['px'])
pltr.plot_values()
pltr.savefig("sliced_muons.png")