Example 03 – Detector uncertainties
Aims
Generate random detector response variations
Include detector uncertainties in the response matrix by using a
ResponseMatrixArrayBuilderInclude statistical uncertainties by generating randomly varied matrices
Use varied response matrices for fits and hypothesis tests
Instructions
The properties of experimental setups are usually only known to a finite precision. The remaining uncertainty on these properties, and thus the uncertainty on the detector response to true events, must be reflected in the response matrix.
We can create an event-by-event variation of the detector response with the provided script:
$ ../simple_experiment/vary_detector.py ../00/modelA_data.txt modelA_data.txt
$ ../simple_experiment/vary_detector.py ../00/modelB_data.txt modelB_data.txt
It creates 100 randomly generated variations of the simulated detector and varies the simulation that was created in example 00 accordingly. Each variation describes one possible real detector and is often called a “toy variation” or a “universe”.
Events are varied in two ways: The values of reconstructed variables can be different for each toy, and each event can get assigned a weight. The former can be used to cover uncertainties in the detector precision and accuracy, while the latter is a simple way to deal with uncertainties in the overall probability of reconstructing an event.
In this case, the values of the reconstructed reco_x are varied and saved
as reco_x_0 to reco_x_99. The detection efficiency is also varied and
the ratio of the events nominal efficiency and the efficiency assuming the toy
detector stored as weight_0 up to weight_99. Let us plot the different
reconstructed distributions we get from the toy variations:
import numpy as np
from remu import binning, plotting
with open("../01/reco-binning.yml") as f:
reco_binning = binning.yaml.full_load(f)
# Get real data
reco_binning.fill_from_csv_file("../00/real_data.txt")
data = reco_binning.get_values_as_ndarray()
# Get toys
n_toys = 100
toyA = []
toyB = []
for i in range(n_toys):
reco_binning.reset()
reco_binning.fill_from_csv_file(
"modelA_data.txt",
weightfield="weight_%i" % (i,),
rename={"reco_x_%i" % (i,): "reco_x"},
buffer_csv_files=True,
)
toyA.append(reco_binning.get_values_as_ndarray())
reco_binning.reset()
reco_binning.fill_from_csv_file(
"modelB_data.txt",
weightfield="weight_%i" % (i,),
rename={"reco_x_%i" % (i,): "reco_x"},
buffer_csv_files=True,
)
toyB.append(reco_binning.get_values_as_ndarray())
# Plot
pltr = plotting.get_plotter(reco_binning)
for A, B in zip(toyA, toyB):
pltr.plot_array(A / 10.0, alpha=0.05, edgecolor="C1", hatch=None)
pltr.plot_array(B / 10.0, alpha=0.05, edgecolor="C2", hatch=None)
A = np.mean(toyA, axis=0)
B = np.mean(toyB, axis=0)
pltr.plot_array(data, label="Data", hatch=None, edgecolor="C0", linewidth=2)
pltr.plot_array(A / 10.0, label="Model A", hatch=None, edgecolor="C1")
pltr.plot_array(B / 10.0, label="Model B", hatch=None, edgecolor="C2")
pltr.legend()
pltr.savefig("data.png")
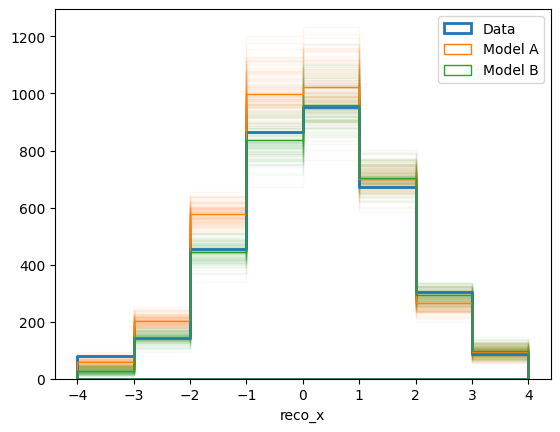
Here we simply plot one histogram for each toy data set with a low alpha value (i.e with high transparency), as well as a solid histogram for the mean values. Also we scaled the model predictions by a factor 10, simply because that makes “experiment running time” of the model predictions correspond to the data.
We are using a few specific arguments to the fill_from_csv_file method:
weightfieldTells the object which field to use as the weight of the events. Does not need to follow any naming conventions. Here we tell it to use the field corresponding to each toy.
renameThe binning of the response matrix expects a variable called
reco_x. This variable is not present in the varied data sets. Here we can specify a dictionary with columns in the data that will be renamed before filling the matrix.buffer_csv_filesReading a CSV file from disk and turning it into an array of floating point values takes quite a bit of time. We can speed up the process by buffering the intermediate array on disk. This means the time consuming parsing of a text file only has to happen once per CSV file.
ReMU handles the detector uncertainties by building a response matrix for each
toy detector separately. These matrices are then used in parallel to calculate
likelihoods. In the end, the marginal (i.e. average) likelihood is used as the
final answer. Some advanced features of ReMU (like sparse matrices, or nuisance
bins) require quite a bit of book-keeping to make the toy matrices behave
consistently and as expected. This is handled by the
ResponseMatrixArrayBuilder:
from remu import binning
from remu import migration
builder = migration.ResponseMatrixArrayBuilder(1)
Its only argument is the number of randomly drawn statistical variations per toy response matrix. The number of simulated events that are used to build the response matrix influences the statistical uncertainty of the response matrix elements. This can be seen as an additional “systematic” uncertainty of the detector response. By generating randomly fluctuated response matrices from the toy matrices, it can be handled organically with the other systematics. In this case we generate 1 randomly varied matrix per toy matrix, yielding a total of 100 matrices in the end.
The toy matrices themselves are built like the nominal matrix in the previous examples and then added to the builder:
with open("../01/reco-binning.yml", 'rt') as f:
reco_binning = binning.yaml.full_load(f)
with open("../01/optimised-truth-binning.yml", 'rt') as f:
truth_binning = binning.yaml.full_load(f)
resp = migration.ResponseMatrix(reco_binning, truth_binning)
n_toys = 100
for i in range(n_toys):
resp.reset()
resp.fill_from_csv_file(["modelA_data.txt", "modelB_data.txt"],
weightfield='weight_%i'%(i,), rename={'reco_x_%i'%(i,): 'reco_x'},
buffer_csv_files=True)
resp.fill_up_truth_from_csv_file(
["../00/modelA_truth.txt", "../00/modelB_truth.txt"],
buffer_csv_files=True)
builder.add_matrix(resp)
Note
The toy variations in modelA_data.txt and modelB_data.txt
must be identical! Otherwise it is not possible to fill the toy matrices with
events from both files. It would mix events reconstructed with different
detectors.
This might create some warnings like this:
UserWarning: Filled-up values are less than the original filling in 1 bins. This should not happen!
return self._replace_smaller_truth(new_truth_binning)
This occurs when events in a bin with already high efficiency are re-weighted to give the impression of an efficiency greater than 100%. If it does not happen too often, it can be ignored.
The builder generates the actual array of floating point numbers as soon as the
ResponseMatrix object is added. It retains no connection to the object
itself.
Now we just need to save the set of response matrices for later use in the likelihood fits:
builder.export("response_matrix.npz")
The LikelihoodCalculator is created just like in the previous example:
import numpy as np
from matplotlib import pyplot as plt
from remu import binning
from remu import plotting
from remu import likelihood
from multiprocess import Pool
pool = Pool(8)
likelihood.mapper = pool.map
with open("../01/reco-binning.yml", 'rt') as f:
reco_binning = binning.yaml.full_load(f)
with open("../01/optimised-truth-binning.yml", 'rt') as f:
truth_binning = binning.yaml.full_load(f)
reco_binning.fill_from_csv_file("../00/real_data.txt")
data = reco_binning.get_entries_as_ndarray()
data_model = likelihood.PoissonData(data)
# No systematics LikelihoodCalculator
response_matrix = "../01/response_matrix.npz"
matrix_predictor = likelihood.ResponseMatrixPredictor(response_matrix)
calc = likelihood.LikelihoodCalculator(data_model, matrix_predictor)
# Systematics LikelihoodCalculator
response_matrix_syst = "response_matrix.npz"
matrix_predictor_syst = likelihood.ResponseMatrixPredictor(response_matrix_syst)
calc_syst = likelihood.LikelihoodCalculator(data_model, matrix_predictor_syst)
To show the influence of the systematic uncertainties, we create two
LikelihoodCalculator objects here. One with the non-varied detector
response and one with the set of systematically varied responses.
Now we can test some models against the data, just like in the previous example:
truth_binning.fill_from_csv_file("../00/modelA_truth.txt")
modelA = truth_binning.get_values_as_ndarray()
modelA /= np.sum(modelA)
truth_binning.reset()
truth_binning.fill_from_csv_file("../00/modelB_truth.txt")
modelB = truth_binning.get_values_as_ndarray()
modelB /= np.sum(modelB)
maxi = likelihood.BasinHoppingMaximizer()
modelA_shape = likelihood.TemplatePredictor([modelA])
calcA = calc.compose(modelA_shape)
retA = maxi(calcA)
print(retA)
fun: 25.257278624558385
log_likelihood: -25.257278624558385
lowest_optimization_result: fun: 25.257278624558385
hess_inv: <1x1 LbfgsInvHessProduct with dtype=float64>
jac: array([8.17123498e-06])
message: 'CONVERGENCE: NORM_OF_PROJECTED_GRADIENT_<=_PGTOL'
nfev: 12
nit: 3
njev: 6
status: 0
success: True
x: array([900.29787236])
message: ['requested number of basinhopping iterations completed successfully']
minimization_failures: 0
nfev: 88
nit: 10
njev: 44
success: True
x: array([900.29787236])
calcA_syst = calc_syst.compose(modelA_shape)
retA_syst = maxi(calcA_syst)
print(retA_syst)
fun: 25.934727545788736
log_likelihood: -25.934727545788736
lowest_optimization_result: fun: 25.934727545788736
hess_inv: <1x1 LbfgsInvHessProduct with dtype=float64>
jac: array([-1.06581326e-06])
message: 'CONVERGENCE: NORM_OF_PROJECTED_GRADIENT_<=_PGTOL'
nfev: 42
nit: 20
njev: 21
status: 0
success: True
x: array([904.96201431])
message: ['requested number of basinhopping iterations completed successfully']
minimization_failures: 0
nfev: 86
nit: 10
njev: 43
success: True
x: array([904.96201431])
modelB_shape = likelihood.TemplatePredictor([modelB])
calcB = calc.compose(modelB_shape)
retB = maxi(calcB)
print(retB)
fun: 24.439068073041028
log_likelihood: -24.439068073041028
lowest_optimization_result: fun: 24.439068073041028
hess_inv: <1x1 LbfgsInvHessProduct with dtype=float64>
jac: array([9.23704824e-06])
message: 'CONVERGENCE: NORM_OF_PROJECTED_GRADIENT_<=_PGTOL'
nfev: 14
nit: 4
njev: 7
status: 0
success: True
x: array([986.50823434])
message: ['requested number of basinhopping iterations completed successfully']
minimization_failures: 0
nfev: 108
nit: 10
njev: 54
success: True
x: array([986.50823434])
calcB_syst = calc_syst.compose(modelB_shape)
retB_syst = maxi(calcB_syst)
print(retB_syst)
fun: 24.948112616354866
log_likelihood: -24.948112616354866
lowest_optimization_result: fun: 24.948112616354866
hess_inv: <1x1 LbfgsInvHessProduct with dtype=float64>
jac: array([3.55271086e-07])
message: 'CONVERGENCE: NORM_OF_PROJECTED_GRADIENT_<=_PGTOL'
nfev: 44
nit: 21
njev: 22
status: 0
success: True
x: array([981.98732246])
message: ['requested number of basinhopping iterations completed successfully']
minimization_failures: 0
nfev: 78
nit: 10
njev: 39
success: True
x: array([981.98732246])
Let us take another look at how the fitted templates and the data compare in reco space. This time we are going to use ReMU’s built in functions to plot a set of bin contents, i.e. the set of model predictions varied by the detector systematics:
pltr = plotting.get_plotter(reco_binning)
pltr.plot_values(edgecolor='C0', label='data', hatch=None, linewidth=2.)
modelA_reco, modelA_weights = calcA.predictor(retA.x)
modelB_reco, modelB_weights = calcB.predictor(retB.x)
modelA_syst_reco, modelA_syst_weights = calcA_syst.predictor(retA.x)
modelB_syst_reco, modelB_syst_weights = calcB_syst.predictor(retB.x)
pltr.plot_array(modelA_reco, label='model A', edgecolor='C1', hatch=None)
pltr.plot_array(modelA_syst_reco, label='model A syst', edgecolor='C1',
hatch=r'//', stack_function=0.68)
pltr.plot_array(modelB_reco, label='model B', edgecolor='C2', hatch=None)
pltr.plot_array(modelB_syst_reco, label='model B syst', edgecolor='C2',
hatch=r'\\', stack_function=0.68)
pltr.legend()
pltr.savefig('reco-comparison.png')
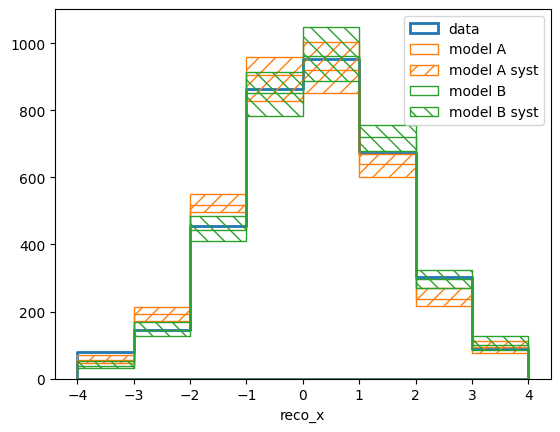
The modelX_syst_reco arrays now contain 100 different predictions,
corresponding to the 100 detector variations. The parameter stac_function ==
0.68 tells the plotter to draw the area of the central 68% of the range of
predictions.
And of course we can compare the p-values of the two fitted models assuming the nominal detector response:
testA = likelihood.HypothesisTester(calcA)
testB = likelihood.HypothesisTester(calcB)
print(testA.max_likelihood_p_value())
print(testB.max_likelihood_p_value())
0.432
0.62
As well as the p-values yielded when taking the systematic uncertainties into account:
testA_syst = likelihood.HypothesisTester(calcA_syst)
testB_syst = likelihood.HypothesisTester(calcB_syst)
print(testA_syst.max_likelihood_p_value())
print(testB_syst.max_likelihood_p_value())
0.556
0.728
We can construct confidence intervals on the template weights of the models using the maximum likelihood p-value just like before:
p_values_A = []
p_values_B = []
p_values_A_syst = []
p_values_B_syst = []
values = np.linspace(600, 1600, 21)
for v in values:
A = testA.max_likelihood_p_value([v])
A_syst = testA_syst.max_likelihood_p_value([v])
B = testB.max_likelihood_p_value([v])
B_syst = testB_syst.max_likelihood_p_value([v])
print(v, A, A_syst, B, B_syst)
p_values_A.append(A)
p_values_B.append(B)
p_values_A_syst.append(A_syst)
p_values_B_syst.append(B_syst)
fig, ax = plt.subplots()
ax.set_xlabel("Model weight")
ax.set_ylabel("p-value")
ax.plot(values, p_values_A, label="Model A", color='C1', linestyle='dotted')
ax.plot(values, p_values_A_syst, label="Model A syst", color='C1', linestyle='solid')
ax.plot(values, p_values_B, label="Model B", color='C2', linestyle='dotted')
ax.plot(values, p_values_B_syst, label="Model B syst", color='C2', linestyle='solid')
ax.axvline(retA.x[0], color='C1', linestyle='dotted')
ax.axvline(retA_syst.x[0], color='C1', linestyle='solid')
ax.axvline(retB.x[0], color='C2', linestyle='dotted')
ax.axvline(retB_syst.x[0], color='C2', linestyle='solid')
ax.axhline(0.32, color='k', linestyle='dashed')
ax.axhline(0.05, color='k', linestyle='dashed')
ax.legend(loc='best')
fig.savefig("p-values.png")
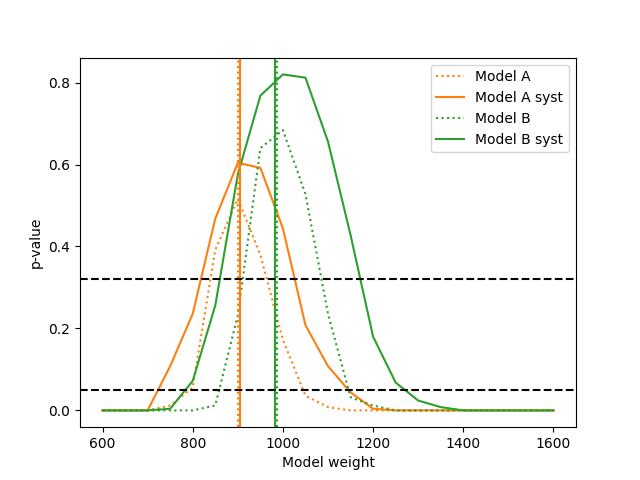
The “fixed” models have no free parameters here, making them degenerate. This
means using the max_likelihood_p_value() is equivalent to testing the
corresponding simple hypotheses using the likelihood_p_value(). No
actual maximisation is taking place. Note that the p-values for model A are
generally smaller than those for model B. This is consistent with previous
results showing that the data is better described by model B.
Finally, let us construct confidence intervals of the template weights, assuming that the corresponding model is correct:
p_values_A = []
p_values_B = []
p_values_A_syst = []
p_values_B_syst = []
values = np.linspace(600, 1600, 21)
for v in values:
A = testA.max_likelihood_ratio_p_value([v])
A_syst = testA_syst.max_likelihood_ratio_p_value([v])
B = testB.max_likelihood_ratio_p_value([v])
B_syst = testB_syst.max_likelihood_ratio_p_value([v])
print(v, A, A_syst, B, B_syst)
p_values_A.append(A)
p_values_B.append(B)
p_values_A_syst.append(A_syst)
p_values_B_syst.append(B_syst)
p_values_A_wilks = []
p_values_B_wilks = []
fine_values = np.linspace(600, 1600, 100)
for v in fine_values:
A = testA_syst.wilks_max_likelihood_ratio_p_value([v])
B = testB_syst.wilks_max_likelihood_ratio_p_value([v])
print(v, A, B)
p_values_A_wilks.append(A)
p_values_B_wilks.append(B)
fig, ax = plt.subplots()
ax.set_xlabel("Model weight")
ax.set_ylabel("p-value")
ax.plot(values, p_values_A, label="Model A", color='C1', linestyle='dotted')
ax.plot(values, p_values_A_syst, label="Model A syst", color='C1', linestyle='solid')
ax.plot(fine_values, p_values_A_wilks, label="Model A Wilks", color='C1', linestyle='dashed')
ax.plot(values, p_values_B, label="Model B", color='C2', linestyle='dotted')
ax.plot(values, p_values_B_syst, label="Model B syst", color='C2', linestyle='solid')
ax.plot(fine_values, p_values_B_wilks, label="Model B Wilks", color='C2', linestyle='dashed')
ax.axvline(retA.x[0], color='C1', linestyle='dotted')
ax.axvline(retA_syst.x[0], color='C1', linestyle='solid')
ax.axvline(retB.x[0], color='C2', linestyle='dotted')
ax.axvline(retB_syst.x[0], color='C2', linestyle='solid')
ax.axhline(0.32, color='k', linestyle='dashed')
ax.axhline(0.05, color='k', linestyle='dashed')
ax.legend(loc='best')
fig.savefig("ratio-p-values.png")
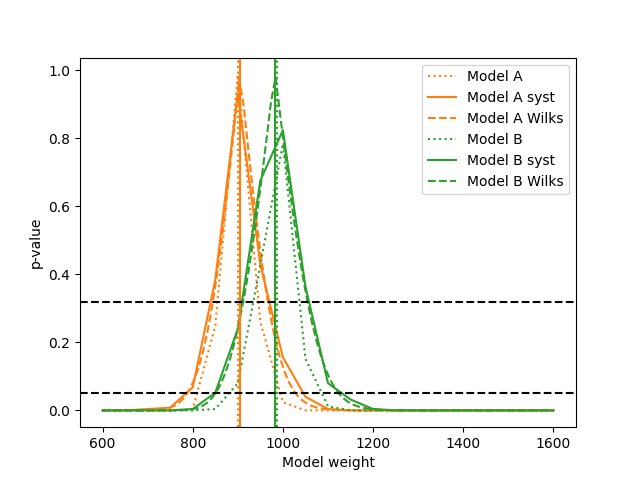
Note that the p-values for both models go up to 1.0 here (within the granularity of the parameter scan) and that the constructed confidence intervals are very different from the ones before. This is because we are asking a different question now. Before we asked the question “What is the probability of getting a worse likelihood assuming that the tested model-parameter is true?”. Now we ask the question “What is the probability of getting a worse best-fit likelihood ratio, assuming the tested model-parameter is true?”. Since the likelihood ratio is 1.0 at the best fit point and the likelihood ratio of nested hypotheses is less than or equal to 1.0 by construction, the p-value is 100% there.
The method wilks_max_likelihood_ratio_p_value() calculates the same
p-value as max_likelihood_ratio_p_value(), but does so assuming Wilks’
theorem holds. This does not require the generation of random data and
subsequent likelihood maximisations, so it is much faster.